| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation date | 2015-10-09 22:33:26 UTC |
|---|
| Update date | 2017-01-19 02:36:41 UTC |
|---|
| FoodComEx ID | PC000923 |
|---|
| FoodDB Record | FDB028319 |
|---|
| Chemical Information |
|---|
| Name | 7-Methyladenine |
|---|
| Description | 7-Methyladenine is part of the base excision repair pathway. In this pathway, alkylated DNA is hydrolysed via the enzyme DNA-3-methyladenine glycosylase II (EC 3.2.2.21), releasing 3-methyladenine, 3-methylguanine, 7-methylguanine and 7-methyladenine. This enzyme is responsible for the hydrolysis of the deoxyribose N-glycosidic bond to excise 3-methyladenine, and 7-methylguanine from the damaged DNA polymer formed by alkylation lesions. (Pathway Commons)
Base excision repair (BER) is a cellular mechanism that repairs damaged DNA throughout the cell cycle. Repairing DNA sequence errors is necessary so that mutations are not propagated or to remove lesions that may lead to breaks in the DNA during replication.
Single bases in DNA can be chemically mutated, for example by deamination or alkylation, resulting in incorrect base-pairing, and consequently, mutations in the DNA. Base excision repair involves flipping the mutated base out of the DNA helix and repairing the base alone. There are two main enzymes used, DNA glycosylases and AP endonucleases. The DNA glycosylase is used to break the beta-N glycosidic bond to create an AP site. AP endonuclease recognizes this site and nicks the damaged DNA on the 5' side (upstream) of the AP site creating a free 3'-OH. DNA polymerase, Pol I (human pol beta), extends the DNA from the free 3'-OH using its exonuclease activity to replace the nucleotide of the damaged base, as well as a few downstream, followed by sealing of the new DNA strand by DNA ligase. In mammalian cells, this is done by LigIII in complex with the scaffold protein XRCC1. Usually, BER is divided into short-patch repair (where a single nucleotide is replaced) or long-patch repair (where 2-10 nucleotides are replaced). Mammalian long-patch repair includes PCNA and pol delta/epsilon for nucleotide resynthesis, FEN1 to cut of the 'flap' including the damaged base, and LigI. [HMDB] |
|---|
| CAS Number | 935-69-3 |
|---|
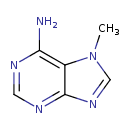
| Structure | |
|---|
| Synonyms | | Synonym | Source |
|---|
| 6-Amino-7-methylpurine | hmdb | | 7-Methyl-7H-purin-6-amine | hmdb | | 7-methyl-7H-purin-6-amine (ACD/Name 4.0) | hmdb | | 7-methyl-7H-purin-6-ylamine (ACD/Name 4.0) | hmdb | | 7-methyl-N7-Methyladenine | hmdb | | 7-Methyladenine | hmdb |
|
|---|
| Chemical Formula | C6H7N5 |
|---|
| IUPAC name | 7-methyl-7H-purin-6-amine |
|---|
| InChI Identifier | InChI=1S/C6H7N5/c1-11-3-10-6-4(11)5(7)8-2-9-6/h2-3H,1H3,(H2,7,8,9) |
|---|
| InChI Key | HCGHYQLFMPXSDU-UHFFFAOYSA-N |
|---|
| Isomeric SMILES | CN1C=NC2=NC=NC(N)=C12 |
|---|
| Average Molecular Weight | 149.1533 |
|---|
| Monoisotopic Molecular Weight | 149.070145249 |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 6-aminopurines. These are purines that carry an amino group at position 6. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | 6-aminopurines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 6-aminopurine
- Aminopyrimidine
- N-substituted imidazole
- Pyrimidine
- Imidolactam
- Azole
- Imidazole
- Heteroaromatic compound
- Azacycle
- Organonitrogen compound
- Hydrocarbon derivative
- Amine
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physico-Chemical Properties - Experimental |
|---|
| Property | Value | Reference |
|---|
| Experimental logP | Not Available | |
|---|
| Experimental Water Solubility | Not Available | |
|---|
| Melting Point | Not Available | |
|---|
|
| Foods of Origin |
|---|
| Food | Content Range | Average | Reference |
|---|
| Food | | | Reference |
|---|
|
| Production Data |
|---|
| Production Method | commercial |
|---|
| Production Method Reference | Not Available |
|---|
| Production Method Reference File | Not Available |
|---|
| Quantity Available | Production upon request, up to 30 mg |
|---|
| Delivery Time | Not Available |
|---|
| Storage Form | solid |
|---|
| Storage Conditions | -80°C |
|---|
| Stability | Not Available |
|---|
| Purity | Not Available |
|---|
| Spectra |
|---|
| Spectral Data Upon Request | Not Available |
|---|
| Provider Information |
|---|
|
| Commercial Vendors |
|---|
| AKSci | 5887AC |
|---|
| Toronto Research Chemicals | M314605 |
|---|